# A tibble: 4 × 3
state year energy
<chr> <dbl> <dbl>
1 DE 2015 8.70
2 IT 2015 9.88
3 DE 2016 8.83
4 IT 2016 10.1 Data preprocessing
Data Visualization and Exploration
File reading
Data formats
There are several formats in which you can find data.
They can be broadly classified in two classes.
Textual data
- human readable
- often easy to parse
- inter-operable in different environments
- slow
- may waste space (but compression helps)
Binary data
- fast
- compact
- requires specialized software to be read
- harder to access in different environments
- not human readable
Comma Separated Values (CSV)
Textual data
One of the simplest textual formats
state, year, energy
DE, 2015, 8.803
IT, 2015, 9.879
DE, 2016, 8.917
IT, 2016, 10.062CSV’s friends
The delimiter separating symbol is not always a comma.
Missing values in the input
Specifying column names
Specifying column names
Data frames
A way of representing data
A data frame is the most common way to represent tabular data in
R.A data frame is essentially a list of vectors.
A data frame has
- several columns with names,
- many rows.
Processing data frames
Usually, you do not manipulate single values directly.
Instead, entire columns are processed all in one go.
tibbles
A tibble is an enhanced data frame with
better printing,
better type handling,
better defaults for building and subsetting,
and possibility to use invalid identifiers as column names.
Our running example
We will use a tibble of all the flights leaving the New Your airport in 2013.
This package contains 5 datasets.
Our running example
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Using dplyr
Dplyr
The
dplyrlibrary is part of thetidyverse.It provides a consistent set of functions to solve most data manipulation problems.
- But I came here for doing visualizations!
- Data rarely comes in a readily usable form.
- This means that we first need to know how to properly “prepare” the data.
General structure of a dplyr call
Usually you will bring all the functions of dplyr in scope using either
or
Filtering rows
# A tibble: 842 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 832 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Filtering rows
- How can we filter all the flights where the delay was less than 10?
- How can we filter all the flights with missing departure delay?
Sorting data
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 12 7 2040 2123 -43 40 2352
2 2013 2 3 2022 2055 -33 2240 2338
3 2013 11 10 1408 1440 -32 1549 1559
4 2013 1 11 1900 1930 -30 2233 2243
5 2013 1 29 1703 1730 -27 1947 1957
6 2013 8 9 729 755 -26 1002 955
7 2013 10 23 1907 1932 -25 2143 2143
8 2013 3 30 2030 2055 -25 2213 2250
9 2013 3 2 1431 1455 -24 1601 1631
10 2013 5 5 934 958 -24 1225 1309
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Sorting data
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 9 641 900 1301 1242 1530
2 2013 6 15 1432 1935 1137 1607 2120
3 2013 1 10 1121 1635 1126 1239 1810
4 2013 9 20 1139 1845 1014 1457 2210
5 2013 7 22 845 1600 1005 1044 1815
6 2013 4 10 1100 1900 960 1342 2211
7 2013 3 17 2321 810 911 135 1020
8 2013 6 27 959 1900 899 1236 2226
9 2013 7 22 2257 759 898 121 1026
10 2013 12 5 756 1700 896 1058 2020
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Getting column names
Selecting columns
Renaming columns
# A tibble: 336,776 × 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ℹ 336,766 more rows
# ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
# tail_num <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
# hour <dbl>, minute <dbl>, time_hour <dttm>Creating new columns
speed_data <- select(flights, distance, air_time)
mutate(speed_data, speed = distance * 60/ air_time)# A tibble: 336,776 × 3
distance air_time speed
<dbl> <dbl> <dbl>
1 1400 227 370.
2 1416 227 374.
3 1089 160 408.
4 1576 183 517.
5 762 116 394.
6 719 150 288.
7 1065 158 404.
8 229 53 259.
9 944 140 405.
10 733 138 319.
# ℹ 336,766 more rowsCreating new columns
How can we add a column with the logarithm of the distance?
Conditional mutation
if_else( condition, value_if_true, value_if_false )# A tibble: 336,776 × 20
dep_delay delayed_str year month day dep_time sched_dep_time arr_time
<dbl> <chr> <int> <int> <int> <int> <int> <int>
1 2 delayed 2013 1 1 517 515 830
2 4 delayed 2013 1 1 533 529 850
3 2 delayed 2013 1 1 542 540 923
4 -1 not_delayed 2013 1 1 544 545 1004
5 -6 not_delayed 2013 1 1 554 600 812
# ℹ 336,771 more rows
# ℹ 12 more variables: sched_arr_time <int>, arr_delay <dbl>, carrier <chr>,
# flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>,
# distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Counting
Summarising
# A tibble: 1 × 1
avg_delay
<dbl>
1 12.6More useful summarisation with group_by
grouped_flights <- group_by(flights, year, month)
summarise(grouped_flights, avg_delay = mean(dep_delay, na.rm = TRUE))# A tibble: 12 × 3
# Groups: year [1]
year month avg_delay
<int> <int> <dbl>
1 2013 1 10.0
2 2013 2 10.8
3 2013 3 13.2
4 2013 4 13.9
5 2013 5 13.0
6 2013 6 20.8
7 2013 7 21.7
8 2013 8 12.6
9 2013 9 6.72
10 2013 10 6.24
11 2013 11 5.44
12 2013 12 16.6 Summarisation
How can we get the maximum and minimum delay by year and month?
# A tibble: 12 × 4
# Groups: year [1]
year month max_delay min_delay
<int> <int> <dbl> <dbl>
1 2013 1 1301 -30
2 2013 2 853 -33
3 2013 3 911 -25
4 2013 4 960 -21
5 2013 5 878 -24
6 2013 6 1137 -21
7 2013 7 1005 -22
8 2013 8 520 -26
9 2013 9 1014 -24
10 2013 10 702 -25
11 2013 11 798 -32
12 2013 12 896 -43Group by
The group_by function produces a grouped tibble.
This means that the operations applied on it act on each group separately.
How can you replicate the behavior of count(flights, month) using group_by, summarise, and n()?
Piping!
Instead of repeatedly assigning the result of each function to a variable that will be used just once, you can use the %>% operator
This operator takes the result of the function on its left
and
makes it the first argument of the function on its right.
Piping example
# A tibble: 12 × 2
month avg_speed
<int> <dbl>
1 1 370.
2 2 375.
3 3 390.
4 4 389.
5 5 407.
6 6 404.
7 7 412.
8 8 408.
9 9 414.
10 10 397.
11 11 385.
12 12 377.Joining two data frames
Joining two data frames
Sometimes the information we need is in different data frames and we need to join them.
This is a family of operations borrowed from the database world.
The basic idea:
we use the values taken by some columns in one table
to match rows in the other table.
Join types
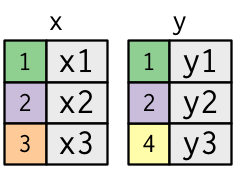
Two columns each: key (coloured) and value (gray)
Join types
# A tibble: 3 × 2
key val_x
<dbl> <chr>
1 1 x1
2 2 x2
3 3 x3 Join types
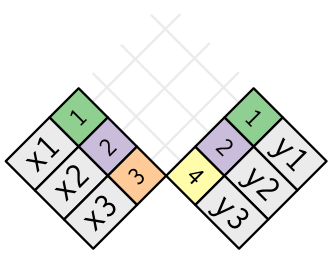
Join types: inner join
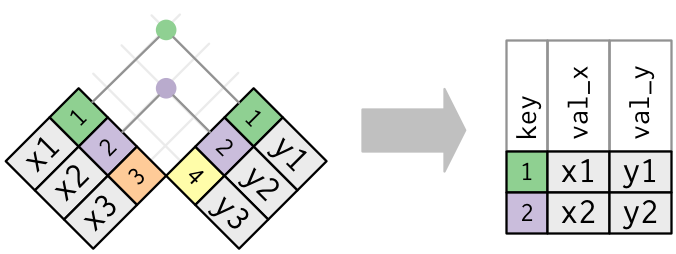
Keep only the rows that have keys in both tables
Join types: outer join
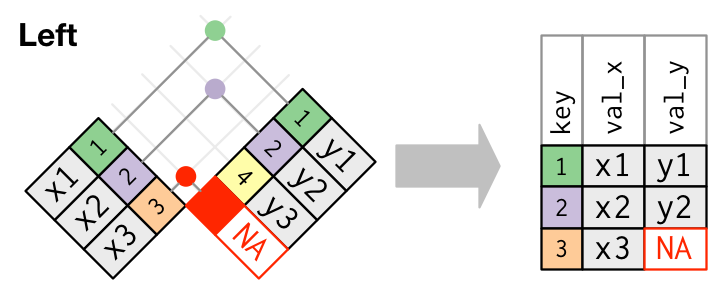
Keep all the rows from left table
Join types: outer join
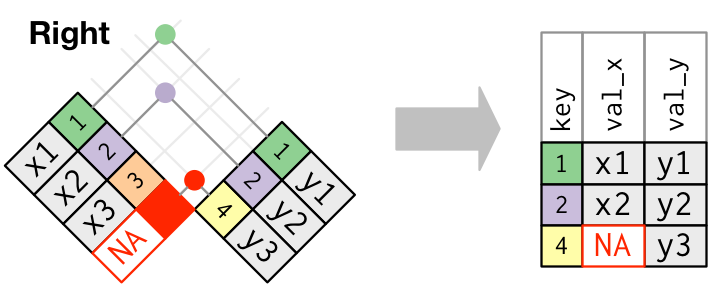
Keep all the rows from right table
Join types: outer join
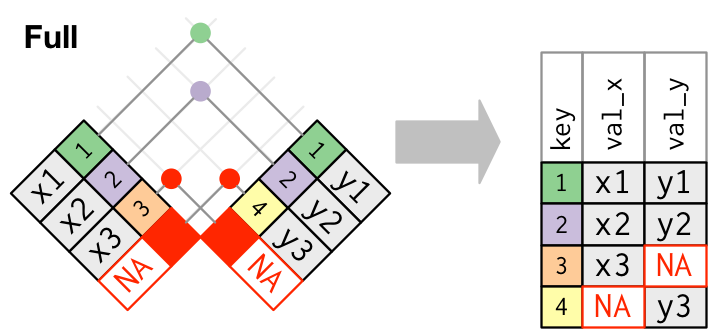
Keep all the rows from both tables
Join: duplicate keys
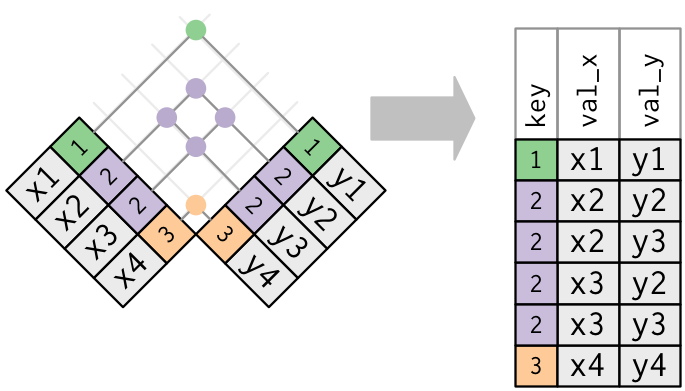
For a given key, get all the possible combinations of values
Join examples
Join examples
Join examples
Join examples
An example on real data
flights2 <- select(flights, year, origin, dest, tailnum)
planes2 <- select(planes, tailnum, year, manufacturer, model)
inner_join(flights2, planes2, by="tailnum")# A tibble: 284,170 × 7
year.x origin dest tailnum year.y manufacturer model
<int> <chr> <chr> <chr> <int> <chr> <chr>
1 2013 EWR IAH N14228 1999 BOEING 737-824
2 2013 LGA IAH N24211 1998 BOEING 737-824
3 2013 JFK MIA N619AA 1990 BOEING 757-223
4 2013 JFK BQN N804JB 2012 AIRBUS A320-232
5 2013 LGA ATL N668DN 1991 BOEING 757-232
6 2013 EWR ORD N39463 2012 BOEING 737-924ER
7 2013 EWR FLL N516JB 2000 AIRBUS INDUSTRIE A320-232
8 2013 LGA IAD N829AS 1998 CANADAIR CL-600-2B19
9 2013 JFK MCO N593JB 2004 AIRBUS A320-232
10 2013 JFK PBI N793JB 2011 AIRBUS A320-232
# ℹ 284,160 more rowsWrap up
Come up with a way to compute the fraction of delayed flights per month.
Tools of the trade
filter
mutate
select
rename
if_else
count
ngroup_by
summarise
inner_join
outer_join
left_join
right_joinSolution 1
all_counts <- flights %>%
filter(!is.na(dep_delay)) %>%
count(month)
delayed_flights <- flights %>%
filter(!is.na(dep_delay)) %>%
filter(dep_delay > 0) %>%
count(month) %>%
rename(n_delayed = n)
inner_join(delayed_flights, all_counts,
by="month") %>%
mutate(
fraction_delayed = n_delayed / n
) %>%
select(month, fraction_delayed)# A tibble: 12 × 2
month fraction_delayed
<int> <dbl>
1 1 0.365
2 2 0.385
3 3 0.401
4 4 0.381
5 5 0.400
6 6 0.465
7 7 0.488
8 8 0.406
9 9 0.288
10 10 0.304
11 11 0.305
12 12 0.500Solution 2
# A tibble: 12 × 2
month fraction_delayed
<int> <dbl>
1 1 0.365
2 2 0.385
3 3 0.401
4 4 0.381
5 5 0.400
6 6 0.465
7 7 0.488
8 8 0.406
9 9 0.288
10 10 0.304
11 11 0.305
12 12 0.500Solution 3
# A tibble: 12 × 2
month fraction_delayed
<int> <dbl>
1 1 0.365
2 2 0.385
3 3 0.401
4 4 0.381
5 5 0.400
6 6 0.465
7 7 0.488
8 8 0.406
9 9 0.288
10 10 0.304
11 11 0.305
12 12 0.500Tidy data
Tidy data
Each variable must have its own column.
Each observation must have its own row.
Each value must have its own cell.
A tidy data frame

Alarm bells
Column names that should be values.
Values that should be column names.
Making data tidy
Most datasets are not tidy
A variable might be spread across multiple columns.
An observation might be scattered across multiple rows.
We mainly have two tools to fix these two problems:
pivot_longer
pivot_wider
Example 1
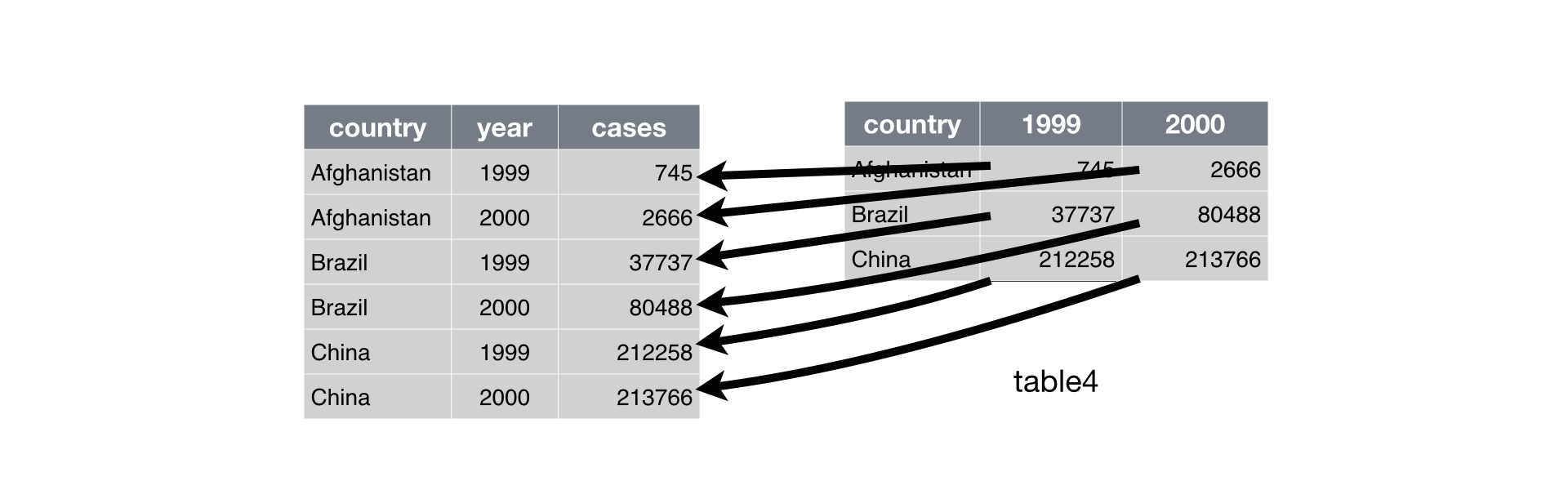
Longer
Was:
# A tibble: 3 × 3
country `1999` `2000`
<chr> <dbl> <dbl>
1 Afghanistan 19987071 20595360
2 Brazil 172006362 174504898
3 China 1272915272 1280428583Longer
Example 2
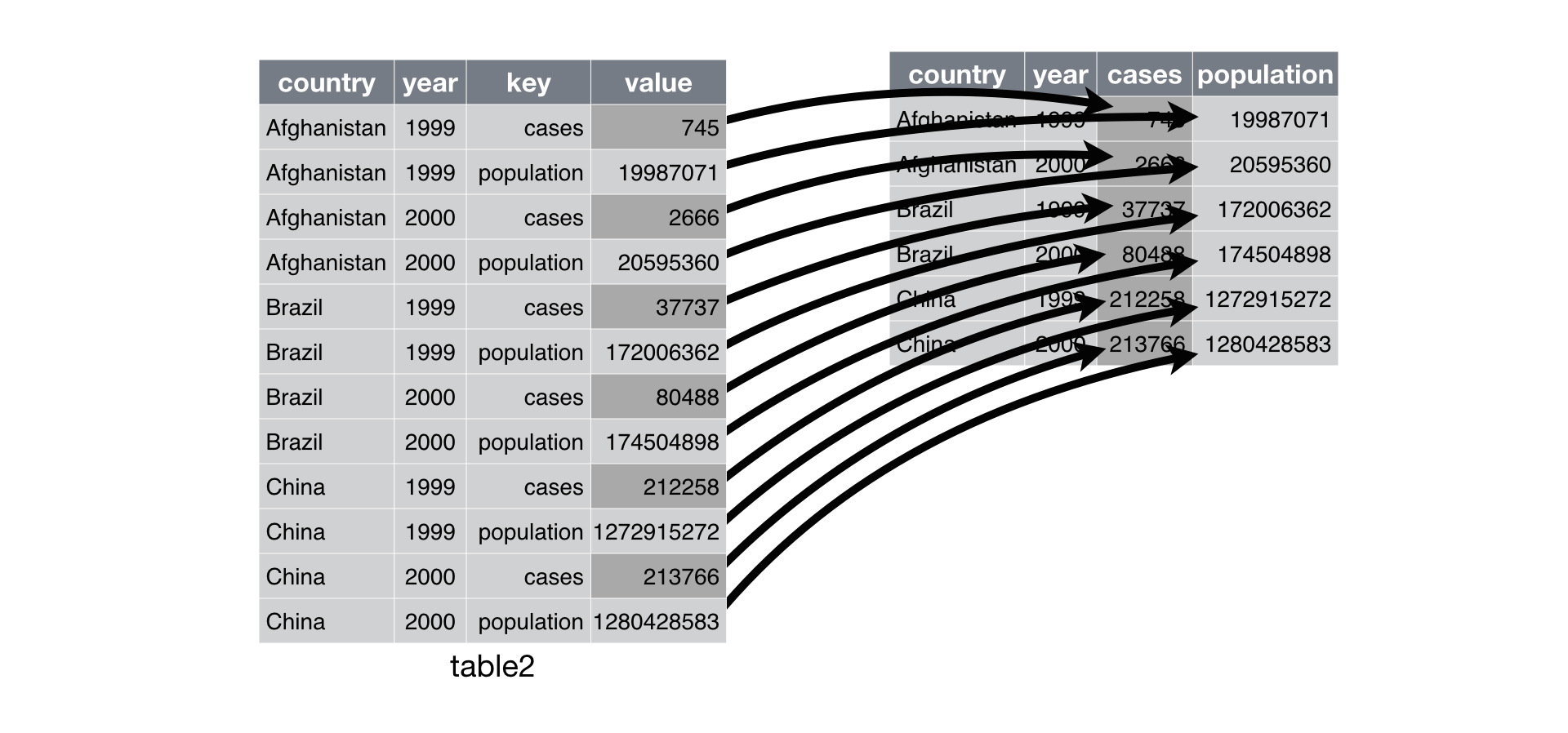
Is this data frame tidy?
# A tibble: 12 × 4
country year type count
<chr> <dbl> <chr> <dbl>
1 Afghanistan 1999 cases 745
2 Afghanistan 1999 population 19987071
3 Afghanistan 2000 cases 2666
4 Afghanistan 2000 population 20595360
5 Brazil 1999 cases 37737
6 Brazil 1999 population 172006362
7 Brazil 2000 cases 80488
8 Brazil 2000 population 174504898
9 China 1999 cases 212258
10 China 1999 population 1272915272
11 China 2000 cases 213766
12 China 2000 population 1280428583An observation might be scattered across multiple rows.
Wider
# A tibble: 6 × 4
country year cases population
<chr> <dbl> <dbl> <dbl>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583
Was:
# A tibble: 12 × 4
country year type count
<chr> <dbl> <chr> <dbl>
1 Afghanistan 1999 cases 745
2 Afghanistan 1999 population 19987071
3 Afghanistan 2000 cases 2666
4 Afghanistan 2000 population 20595360
5 Brazil 1999 cases 37737
6 Brazil 1999 population 172006362
7 Brazil 2000 cases 80488
8 Brazil 2000 population 174504898
9 China 1999 cases 212258
10 China 1999 population 1272915272
11 China 2000 cases 213766
12 China 2000 population 1280428583Wider
Example 3
Is this table tidy?
Separating
# A tibble: 6 × 4
country year cases population
<chr> <dbl> <chr> <chr>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583Is there something odd?
Separating
# A tibble: 6 × 4
country year cases population
<chr> <dbl> <int> <int>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583Separating
# A tibble: 6 × 4
country year cases population
<chr> <dbl> <int> <int>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583A real-world example
Available from https://ec.europa.eu/eurostat/api/dissemination/sdmx/2.1/data/educ_uoe_grad05?format=tsv
freq,unit,isced11,age,geo\TIME_PERIOD 2013 2014 2015 2016 2017 2018 2019 2020 2021 2022
A,P_THAB,ED5,TOTAL,AL : : : : : : : : 2.9 3.8
A,P_THAB,ED5,TOTAL,AT 25.1 24.8 24.3 21.8 22.7 22.8 22.7 25.5 23.8 22.3
A,P_THAB,ED5,TOTAL,BE 1.8 1.8 : d 2.1 2.1 2.0 2.0 d 3.8 d 5.1 4.9
A,P_THAB,ED5,TOTAL,BG : z : z : z : z : z : z : z : z : z : z
A,P_THAB,ED5,TOTAL,CH 2.2 1.9 0.5 0.4 0.3 0.3 0.3 0.3 0.4 :
A,P_THAB,ED5,TOTAL,CY 7.3 7.5 6.9 7.0 6.4 7.8 7.1 7.4 9.1 7.8
A,P_THAB,ED5,TOTAL,CZ 0.3 0.3 0.3 0.3 0.3 0.3 0.3 0.3 0.4 0.4 Discuss how to load this file into a tidy dataframe.
read_tsv(path, na, ....)
pivot_longer(data, columns, names_to=key, values_to=value)
pivot_wider(data, names_from=key, values_from=value)
separate(data, col, into, ...)Cheat sheets
A real world example
# A tibble: 78 × 21
`unit,geo\\time` `2000` `2001` `2002` `2003` `2004` `2005` `2006` `2007`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 EUR_KGOE,AT 8.69 8.34 8.43 8.08 8.15 8.06 8.26 8.72
2 EUR_KGOE,BA NA NA NA NA NA NA NA NA
3 EUR_KGOE,BE 4.73 4.83 4.95 4.77 4.91 5.05 5.18 5.38
4 EUR_KGOE,BG 1.32 1.31 1.42 1.45 1.59 1.61 1.67 1.81
# ℹ 74 more rows
# ℹ 12 more variables: `2008` <dbl>, `2009` <dbl>, `2010` <dbl>, `2011` <dbl>,
# `2012` <dbl>, `2013` <dbl>, `2014` <dbl>, `2015` <dbl>, `2016` <dbl>,
# `2017` <dbl>, `2018` <dbl>, `2019` <dbl>energy_productivity %>%
pivot_longer(`2000`:`2019`, names_to='year', values_to='energy_productivity')# A tibble: 1,560 × 3
`unit,geo\\time` year energy_productivity
<chr> <chr> <dbl>
1 EUR_KGOE,AT 2000 8.69
2 EUR_KGOE,AT 2001 8.34
3 EUR_KGOE,AT 2002 8.43
4 EUR_KGOE,AT 2003 8.08
5 EUR_KGOE,AT 2004 8.15
6 EUR_KGOE,AT 2005 8.06
7 EUR_KGOE,AT 2006 8.26
8 EUR_KGOE,AT 2007 8.72
9 EUR_KGOE,AT 2008 8.78
10 EUR_KGOE,AT 2009 8.90
# ℹ 1,550 more rowsenergy_productivity %>%
pivot_longer(`2000`:`2019`, names_to='year', values_to='energy_productivity') %>%
separate(`unit,geo\\time`, into=c("unit", "state"), sep=',')# A tibble: 1,560 × 4
unit state year energy_productivity
<chr> <chr> <chr> <dbl>
1 EUR_KGOE AT 2000 8.69
2 EUR_KGOE AT 2001 8.34
3 EUR_KGOE AT 2002 8.43
4 EUR_KGOE AT 2003 8.08
5 EUR_KGOE AT 2004 8.15
6 EUR_KGOE AT 2005 8.06
7 EUR_KGOE AT 2006 8.26
8 EUR_KGOE AT 2007 8.72
9 EUR_KGOE AT 2008 8.78
10 EUR_KGOE AT 2009 8.90
# ℹ 1,550 more rowsBonus: finding problematic values
If you have a dataframe with NA values, you can list them using the following snippet.
# This is the same dataset we used in the assignment
url_education <- "https://ec.europa.eu/eurostat/api/dissemination/sdmx/2.1/data/educ_uoe_grad05?format=tsv"
# We load it from the file, dealing with the NA encodings we already know of
education_raw <- read_tsv(url_education,na=c(":",": z",": d")) %>%
mutate(across(everything(), as.character))
education_raw %>%
# Then we pivot the columns for more convenient processing
pivot_longer(`2022`:`2013`, names_to="year", values_to="value") %>%
# Once you find the problematic parts of the strings, you can uncomment the
# following line to remove the problematic characters
# mutate(value = str_remove(value, "d")) %>%
# Here we keep only the values whose parsing results in a NA
filter(is.na(as.double(value))) %>%
# With distinct you remove the duplicates, getting an overview of the problematic values
distinct(value)# A tibble: 147 × 1
value
<chr>
1 <NA>
2 3.8 d
3 2.0 d
4 8.6 d
5 7.7 d
6 9.3 d
7 8.0 d
8 21.9 d
9 27.1 d
10 22.8 d
# ℹ 137 more rowsData Visualization and Exploration - Data preprocessing - ozan-k.com